|
|||||||||||||||||
|
|||||||||||||||||
|
Research
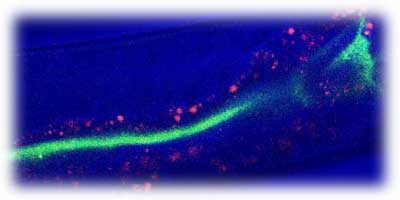
Our laboratory is now divided 50/50 between the Arabidopsis and C. elegans systems. The rationale is that there is a growing body of evidence demonstrating that many of the underlying mechanisms of microbial pathogenesis and corresponding host defense responses are similar in plants and animals and that the study of Arabidopsis and C. elegans defense responses will be synergistic. One example of how bacterial virulence factors are similar in plant and animal pathogens is the conservation of so-called type III protein secretory mechanisms that are involved in the translocation of proteinaceous virulence factors into host cells. Another example is our discovery that Pseudomonas aeruginosa utilizes a shared subset of virulence factors to infect both plants and animals. The use of experimentally tractable model hosts allows us to perform genetics on both sides of the host-pathogen interaction. To better understand the suite of bacterial virulence factors that are used to inflict damage on the host and to circumvent the host’s defense responses, we are developing genomic tools for a pathogenic isolate of P. aeruginosa, strain PA14, which is extremely virulent in both the Arabidopsis and C. elegans models. A full-genome, non-redundant library of defined transposon insertions will allow for high-throughput, saturating screens to identify novel virulence factors, and candidate strains will be selected for studies utilizing combinatorial genetics between sets of host and pathogen mutants.
|
|||||||||||||||||
|
|
|||||||||||||||||
| Copyright © 2006-2012 The Massachusetts General Hospital | |||